library(ggplot2)
library(dplyr)
seed <- 20230327Rで回帰分析:ロバスト回帰
Rでデータサイエンス
ロバスト回帰
サンプルデータの作成
# サンプルデータの作成
set.seed(seed = seed)
n <- 10
x <- runif(n = n, min = 0, max = 10) %>%
sort()
b0 <- 5
b1 <- 10
y <- x * b1 + b0 + rnorm(n = n)
y[n] <- y[n] * 5
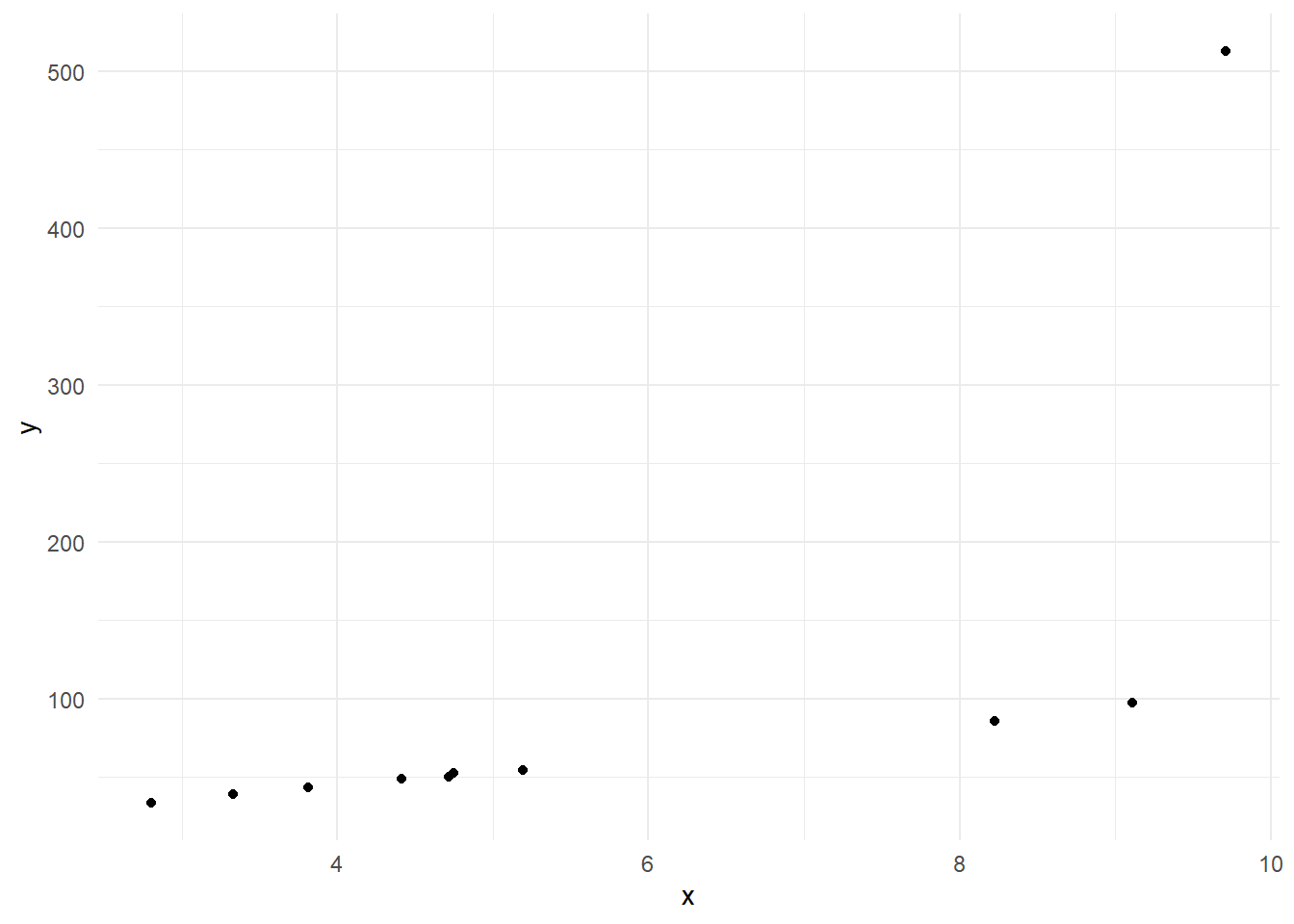
(g <- ggplot(data = data.frame(x, y), mapping = aes(x = x, y = y)) + geom_point() + theme_minimal())# 線形回帰
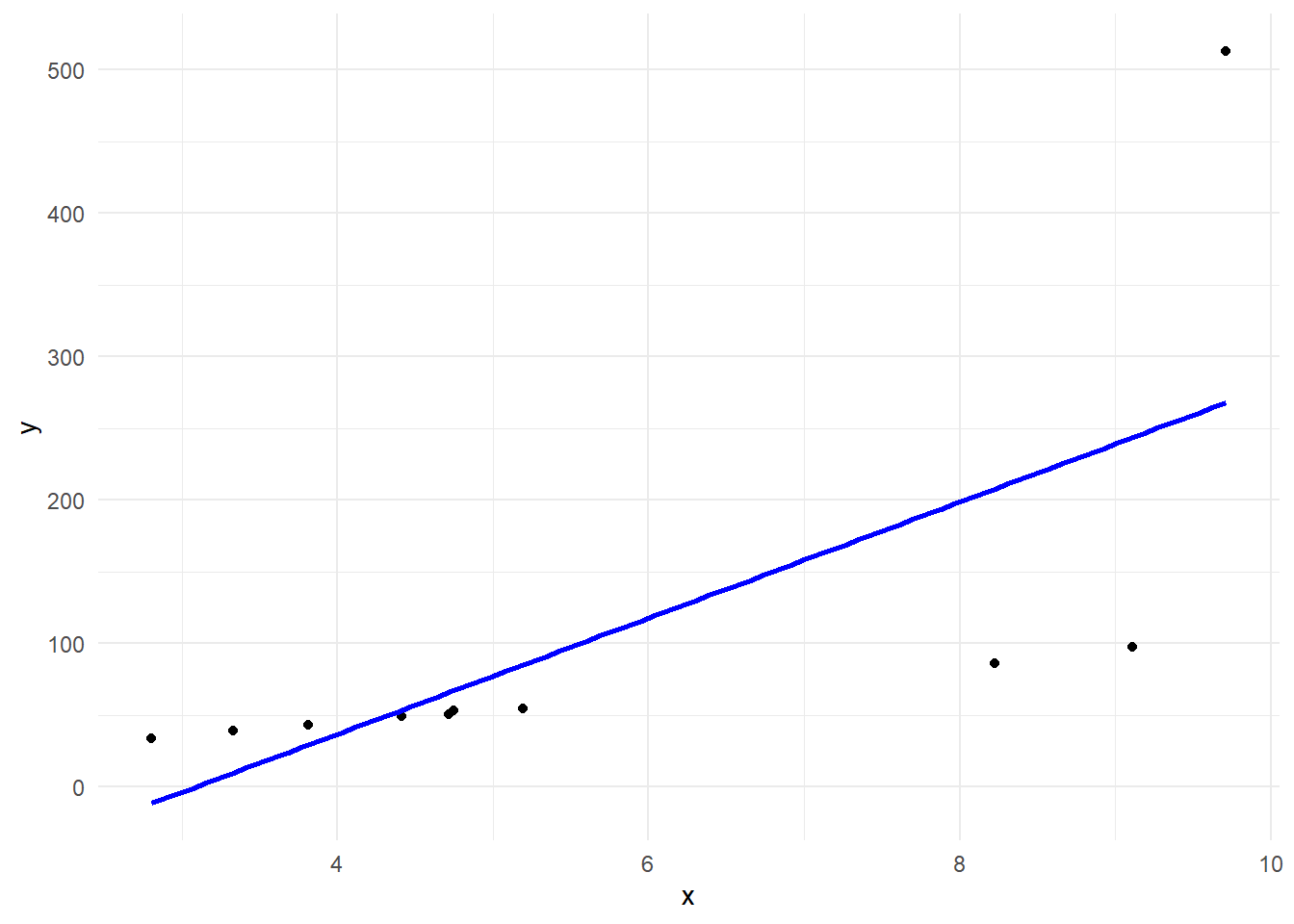
(g <- g + geom_smooth(method = "lm", se = F, col = "blue"))
lm(y ~ x) %>%
summary()
Call:
lm(formula = y ~ x)
Residuals:
Min 1Q Median 3Q Max
-146.421 -26.938 -9.357 25.520 244.812
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -124.82 91.64 -1.362 0.2103
x 40.43 15.07 2.682 0.0278 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 112.2 on 8 degrees of freedom
Multiple R-squared: 0.4735, Adjusted R-squared: 0.4077
F-statistic: 7.195 on 1 and 8 DF, p-value: 0.02782rlm {MASS} によるロバスト線形回帰
# ロバスト線形回帰
library(MASS)
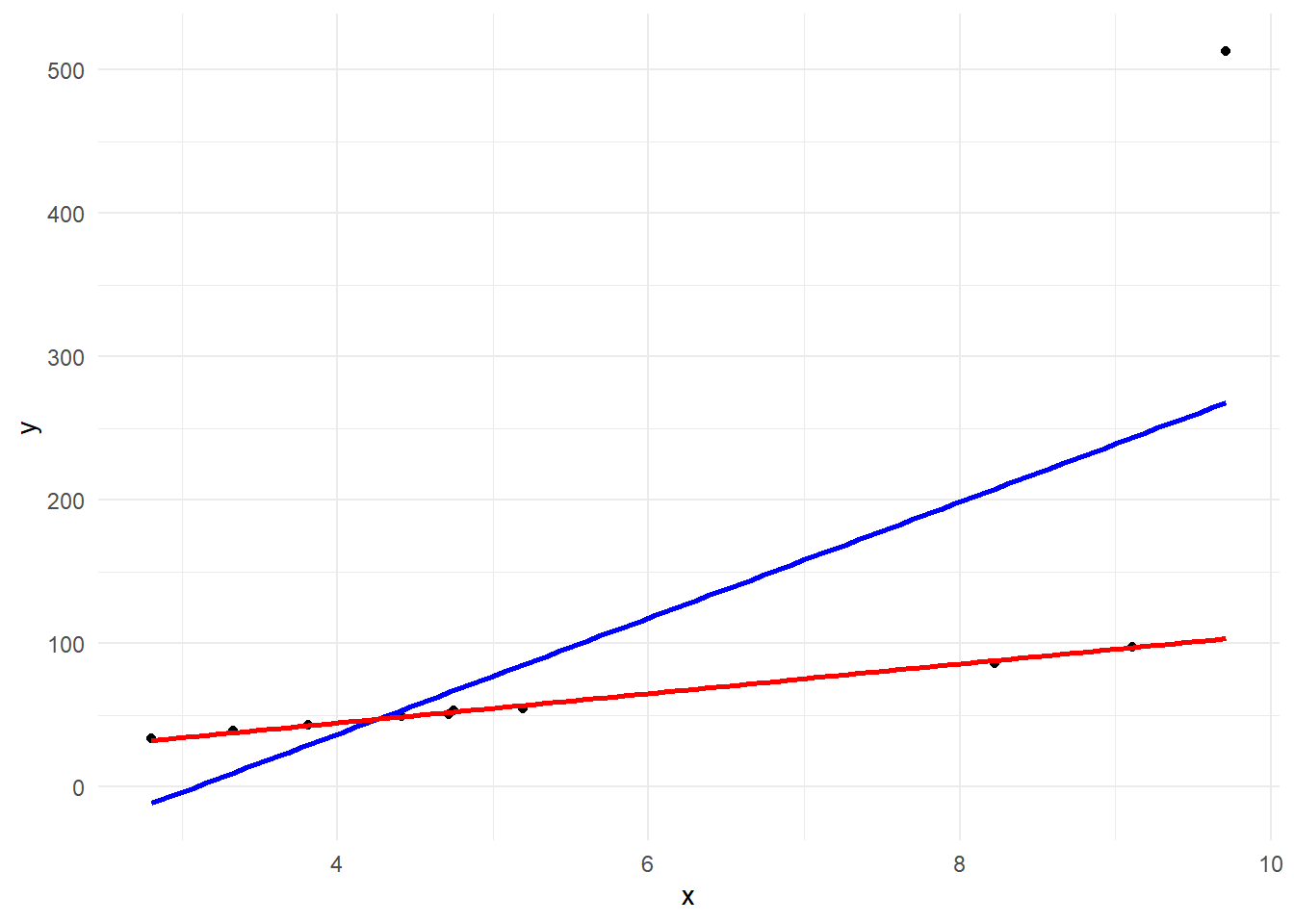
packageVersion("MASS")[1] '7.3.60.0.1'g + geom_smooth(method = "rlm", se = F, col = "red")
result <- rlm(y ~ x)
result %>%
summary()
Call: rlm(formula = y ~ x)
Residuals:
Min 1Q Median 3Q Max
-2.5882 -1.2076 0.2692 0.9066 409.3434
Coefficients:
Value Std. Error t value
(Intercept) 3.6571 1.7619 2.0757
x 10.2574 0.2898 35.3996
Residual standard error: 1.779 on 8 degrees of freedomrlm {MASS}
- 繰返し加重最小二乗(IRLS: Iteratively Reweighted Least Squares)によるロバスト回帰。
- M推定とMM推定から選択。
- \(\psi\,\)関数(影響関数)のデフォルトはHuber。Hampelおよびbisquare(Tukey)も可能。但しMM推定の場合はbisquare(Tukey)。
- 尺度パラメータ(\(s\))はMAD(中央絶対偏差)、Huberおよびproposal 2(Huber’s proposal 2)から選択。
methods("rlm")[1] rlm.default* rlm.formula*
see '?methods' for accessing help and source coderlm.formula
getS3method(f = "rlm", class = "formula")function (formula, data, weights, ..., subset, na.action, method = c("M",
"MM", "model.frame"), wt.method = c("inv.var", "case"), model = TRUE,
x.ret = TRUE, y.ret = FALSE, contrasts = NULL)
{
mf <- match.call(expand.dots = FALSE)
mf$method <- mf$wt.method <- mf$model <- mf$x.ret <- mf$y.ret <- mf$contrasts <- mf$... <- NULL
mf[[1L]] <- quote(stats::model.frame)
mf <- eval.parent(mf)
method <- match.arg(method)
wt.method <- match.arg(wt.method)
if (method == "model.frame")
return(mf)
mt <- attr(mf, "terms")
y <- model.response(mf)
offset <- model.offset(mf)
if (!is.null(offset))
y <- y - offset
x <- model.matrix(mt, mf, contrasts)
xvars <- as.character(attr(mt, "variables"))[-1L]
if ((yvar <- attr(mt, "response")) > 0L)
xvars <- xvars[-yvar]
xlev <- if (length(xvars) > 0L) {
xlev <- lapply(mf[xvars], levels)
xlev[!sapply(xlev, is.null)]
}
weights <- model.weights(mf)
if (!length(weights))
weights <- rep(1, nrow(x))
fit <- rlm.default(x, y, weights, method = method, wt.method = wt.method,
...)
fit$terms <- mt
cl <- match.call()
cl[[1L]] <- as.name("rlm")
fit$call <- cl
fit$contrasts <- attr(x, "contrasts")
fit$xlevels <- .getXlevels(mt, mf)
fit$na.action <- attr(mf, "na.action")
if (model)
fit$model <- mf
if (!x.ret)
fit$x <- NULL
if (y.ret)
fit$y <- y
fit$offset <- offset
if (!is.null(offset))
fit$fitted.values <- fit$fitted.values + offset
fit
}
<bytecode: 0x0000022eee84d5e0>
<environment: namespace:MASS>rlm.default
getS3method(f = "rlm", class = "default")function (x, y, weights, ..., w = rep(1, nrow(x)), init = "ls",
psi = psi.huber, scale.est = c("MAD", "Huber", "proposal 2"),
k2 = 1.345, method = c("M", "MM"), wt.method = c("inv.var",
"case"), maxit = 20, acc = 0.0001, test.vec = "resid",
lqs.control = NULL)
{
irls.delta <- function(old, new) sqrt(sum((old - new)^2)/max(0.00000000000000000001,
sum(old^2)))
irls.rrxwr <- function(x, w, r) {
w <- sqrt(w)
max(abs((matrix(r * w, 1L, length(r)) %*% x)/sqrt(matrix(w,
1L, length(r)) %*% (x^2))))/sqrt(sum(w * r^2))
}
wmad <- function(x, w) {
o <- sort.list(abs(x))
x <- abs(x)[o]
w <- w[o]
p <- cumsum(w)/sum(w)
n <- sum(p < 0.5)
if (p[n + 1L] > 0.5)
x[n + 1L]/0.6745
else (x[n + 1L] + x[n + 2L])/(2 * 0.6745)
}
method <- match.arg(method)
wt.method <- match.arg(wt.method)
nmx <- deparse(substitute(x))
if (is.null(dim(x))) {
x <- as.matrix(x)
colnames(x) <- nmx
}
else x <- as.matrix(x)
if (is.null(colnames(x)))
colnames(x) <- paste("X", seq(ncol(x)), sep = "")
if (qr(x)$rank < ncol(x))
stop("'x' is singular: singular fits are not implemented in 'rlm'")
if (!(any(test.vec == c("resid", "coef", "w", "NULL")) ||
is.null(test.vec)))
stop("invalid 'test.vec'")
xx <- x
yy <- y
if (!missing(weights)) {
if (length(weights) != nrow(x))
stop("length of 'weights' must equal number of observations")
if (any(weights < 0))
stop("negative 'weights' value")
if (wt.method == "inv.var") {
fac <- sqrt(weights)
y <- y * fac
x <- x * fac
wt <- NULL
}
else {
w <- w * weights
wt <- weights
}
}
else wt <- NULL
if (method == "M") {
scale.est <- match.arg(scale.est)
if (!is.function(psi))
psi <- get(psi, mode = "function")
arguments <- list(...)
if (length(arguments)) {
pm <- pmatch(names(arguments), names(formals(psi)),
nomatch = 0L)
if (any(pm == 0L))
warning("some of ... do not match")
pm <- names(arguments)[pm > 0L]
formals(psi)[pm] <- unlist(arguments[pm])
}
if (is.character(init)) {
temp <- if (init == "ls")
lm.wfit(x, y, w, method = "qr")
else if (init == "lts") {
if (is.null(lqs.control))
lqs.control <- list(nsamp = 200L)
do.call("lqs", c(list(x, y, intercept = FALSE),
lqs.control))
}
else stop("'init' method is unknown")
coef <- temp$coefficients
resid <- temp$residuals
}
else {
if (is.list(init))
coef <- init$coef
else coef <- init
resid <- drop(y - x %*% coef)
}
}
else if (method == "MM") {
scale.est <- "MM"
temp <- do.call("lqs", c(list(x, y, intercept = FALSE,
method = "S", k0 = 1.548), lqs.control))
coef <- temp$coefficients
resid <- temp$residuals
psi <- psi.bisquare
if (length(arguments <- list(...)))
if (match("c", names(arguments), nomatch = 0L)) {
c0 <- arguments$c
if (c0 > 1.548)
formals(psi)$c <- c0
else warning("'c' must be at least 1.548 and has been ignored")
}
scale <- temp$scale
}
else stop("'method' is unknown")
done <- FALSE
conv <- NULL
n1 <- (if (is.null(wt))
nrow(x)
else sum(wt)) - ncol(x)
theta <- 2 * pnorm(k2) - 1
gamma <- theta + k2^2 * (1 - theta) - 2 * k2 * dnorm(k2)
if (scale.est != "MM")
scale <- if (is.null(wt))
mad(resid, 0)
else wmad(resid, wt)
for (iiter in 1L:maxit) {
if (!is.null(test.vec))
testpv <- get(test.vec)
if (scale.est != "MM") {
scale <- if (scale.est == "MAD")
if (is.null(wt))
median(abs(resid))/0.6745
else wmad(resid, wt)
else if (is.null(wt))
sqrt(sum(pmin(resid^2, (k2 * scale)^2))/(n1 *
gamma))
else sqrt(sum(wt * pmin(resid^2, (k2 * scale)^2))/(n1 *
gamma))
if (scale == 0) {
done <- TRUE
break
}
}
w <- psi(resid/scale)
if (!is.null(wt))
w <- w * weights
temp <- lm.wfit(x, y, w, method = "qr")
coef <- temp$coefficients
resid <- temp$residuals
if (!is.null(test.vec))
convi <- irls.delta(testpv, get(test.vec))
else convi <- irls.rrxwr(x, w, resid)
conv <- c(conv, convi)
done <- (convi <= acc)
if (done)
break
}
if (!done)
warning(gettextf("'rlm' failed to converge in %d steps",
maxit), domain = NA)
fitted <- drop(xx %*% coef)
cl <- match.call()
cl[[1L]] <- as.name("rlm")
fit <- list(coefficients = coef, residuals = yy - fitted,
wresid = resid, effects = temp$effects, rank = temp$rank,
fitted.values = fitted, assign = temp$assign, qr = temp$qr,
df.residual = NA, w = w, s = scale, psi = psi, k2 = k2,
weights = if (!missing(weights)) weights, conv = conv,
converged = done, x = xx, call = cl)
class(fit) <- c("rlm", "lm")
fit
}
<bytecode: 0x0000022eee89c7f0>
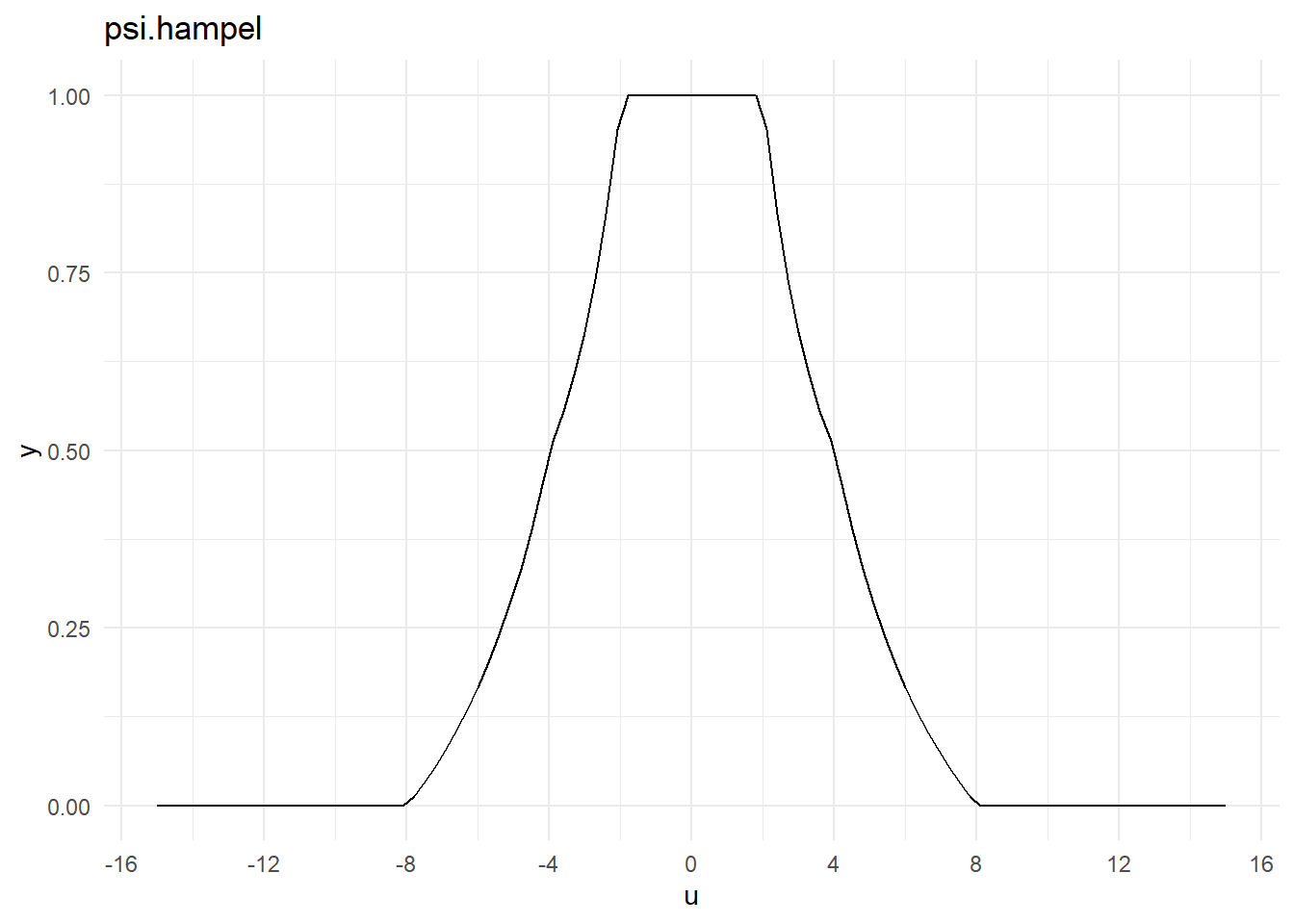
<environment: namespace:MASS>\(\psi\)関数
psi.hampel
psi.hampelfunction (u, a = 2, b = 4, c = 8, deriv = 0)
{
U <- pmin(abs(u) + 0.00000000000000000000000000000000000000000000000001,
c)
if (!deriv)
return(ifelse(U <= a, U, ifelse(U <= b, a, a * (c - U)/(c -
b)))/U)
ifelse(abs(u) <= c, ifelse(U <= a, 1, ifelse(U <= b, 0, -a/(c -
b))), 0)
}
<bytecode: 0x0000022ecc9522c0>
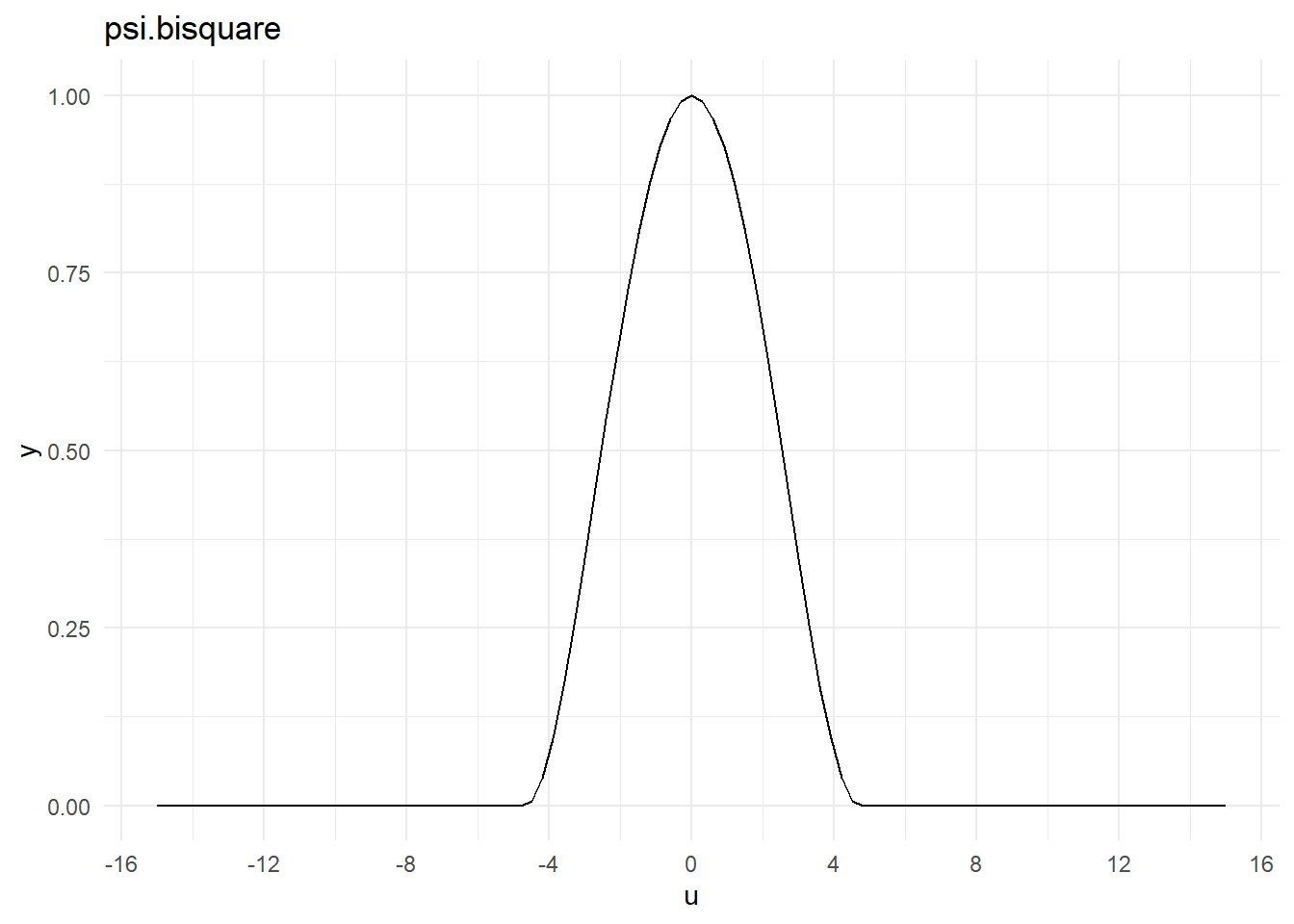
<environment: namespace:MASS>psi.bisquare
psi.bisquarefunction (u, c = 4.685, deriv = 0)
{
if (!deriv)
return((1 - pmin(1, abs(u/c))^2)^2)
t <- (u/c)^2
ifelse(t < 1, (1 - t) * (1 - 5 * t), 0)
}
<bytecode: 0x0000022eedf8b0b0>
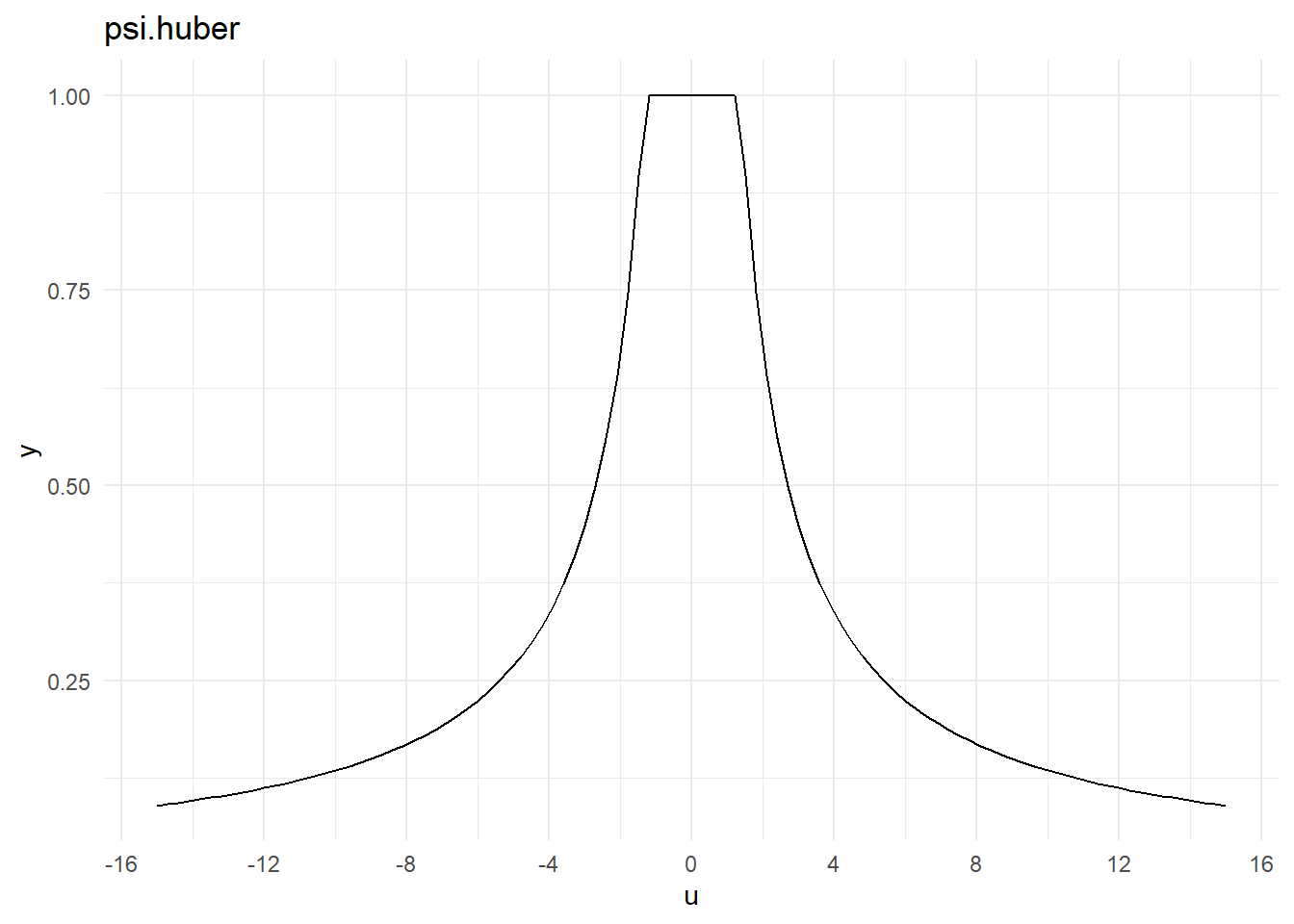
<environment: namespace:MASS>psi.huber
psi.huberfunction (u, k = 1.345, deriv = 0)
{
if (!deriv)
return(pmin(1, k/abs(u)))
abs(u) <= k
}
<bytecode: 0x0000022eee8c7050>
<environment: namespace:MASS>参考引用資料
- https://rdrr.io/cran/MASS/man/rlm.html
- https://www.stat.go.jp/training/2kenkyu/ihou/76/pdf/2-2-767.pdf
- https://jp.mathworks.com/help/stats/robustfit.html
- https://qiita.com/ssugasawa/items/d1a015bff9ee2c8552de
- https://www.stats.ox.ac.uk/~ripley/StatMethods/Robust.pdf
- https://www.stat.go.jp/training/2kenkyu/ihou/69/pdf/2-2-692.pdf
最終更新
Sys.time()[1] "2024-04-03 04:38:02 JST"R、Quarto、Package
R.Version()$version.string[1] "R version 4.3.3 (2024-02-29 ucrt)"quarto::quarto_version()[1] '1.4.542'packageVersion(pkg = "tidyverse")[1] '2.0.0'